Serna Bio Polaris GenAI Platform
After working at PTC Therapeutics, I began working at Serna Bio. They were, surprisingly enough, drawn to my “code-review” background. I would work for Serna Bio a total of three times as of April 2025. The first time was a three month contract that had me reviewing existing and new code from over ten repos. In addition, I would consolidate code from across these various repos into a singular repo that would standardize, test, and add additional functionality to common or fundamental operations. This utilities package would be my primary addition to the company across my initial three month contract.
A month after my initial contract ended, I got another call from the team offering me a full-time position as a member of their ChemAI team. I accepted, and took on a whole new set of responsiblities. Though my original duties continued, I was also tasked with standardizing code for the development of multi-dimensional chemical-space visualizations, and running this code to create new visualizations upon request. In addition, I would make a similar code-based utilities package for the bioinformatics team (this time in R rather than Python). After a few months, I was in charge of planning and facilitating regular meetings regarding IT issues and coding standards. Finally, I was the main point person put in charge of evaluating potential external vendors who would provide a data infrastructure tool that supported the changing data needs of the company. As part of a small start-up, my role was demanding, but it built my communication skills, data infrastructure experience, AWS knowledge, and coding capabilities.
As is true for all start-ups, the priorities of the company eventually shifted, and my work was no longer needed at the company for some time…
Until they called back a few months later asking if I was available for another bit of contract work. This work revolved around developing and dockerizing multiple pipelines for deployment. My primary work would be on the backend of their new tool, Polaris, with Anton Petrov working on the frontend.
The tool and its usage are documented in a blog post made by the company, but basically, the tool uses various proprietary models to generate compounds that are chemically similar to an input seed compound. A mixture of proprietary and non-proprietary models are used to evaluate all of the molecules generated. These metrics are then pareto optimized in order to assign each compound a rank. All of this is displayed visually (thank you Anton), but the underlying data can also be downloaded for programatic evaluation.
During this final contract, I also standardized and simplified other pipelines as part of my work, but as this work is not part of a public blog post, I am unable to discuss the nature of these pipelines.
Each of my stints at Serna taught me different skills and had me working on diverse projects. I appreciate all that I have learned as both a software developer and a start-up member from my time there. I would also like to thank my manager for all three stints, Tim Allen. Tim was supportive and reassuring when I had feelings of Imposter Syndrome at a company with so many skilled and hardworking teammates.
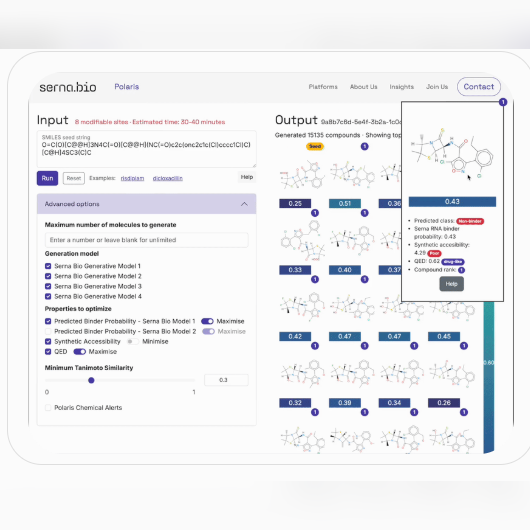
For more on Polaris, see Serna’s blog post here